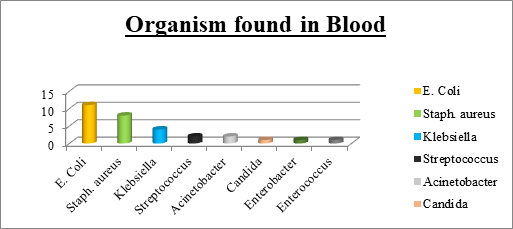
Bacterial bloodstream infections are important causes of morbidity and mortality, globally. The aim of the present study was to determine the bacterial profile of bloodstream infections and their antibiotic susceptibility pattern among the patients admitted to ICU at a tertiary care hospital.This prospective study was conducted over a period of eighteen months. Inclusion criteria comprised of patients admitted to ICU who belonged to either gender and were in the age group of 15-60 years. Over the course of study, 30 out of total 140 blood culture samples were identified to be culture positive (18 GNB and 11GPB). The most common Gram-positive isolate was Staphylococcus spp (26%) while Escherichia coli was the most common gram negative isolate (36%).Escherichia coli expressed highest resistance to all the drugs but sensitivity to Meropenemand Polymyxin B was 72% and 90%, respectively. High degree of resistance was noted to cephalosporins and piperacillin -tazobactam, among all the groups. The study indicated high level of antimicrobial resistance among Gram negative bacilli, esp E.Coli and justifies the need for antimicrobial stewardship to prevent development of further resistance.
Bloodstream infection is one of the principal causes of morbidity and mortality in the intensive care unit. Critical care patients are often associated with an increasing number of invasive devices and monitors that make them five to seven-fold more susceptible to acquisition of nosocomial infection as compared to general inpatients in the hospital [1, 2]. The Surviving sepsis campaign guidelines, ever since its inception, have emphasised the initiation of antibacterial therapy within the first hour of presentation to the hospital for better/improved survival [3]. But, the institution of an inappropriate empirical antimicrobial therapy has been associated with a five-fold reduction in survival. Rapid and accurate identification of bacterial species in the blood is, therefore, of paramount importance [4].
Since, microbiological culture results are not available until after 24 to 72 hours, the initial therapy for infection is often empirical and guided by the clinical presentation. Broad-spectrum antimicrobial agents are generally started initially with the intent to cover most pathogens commonly associated with specific clinical syndromes. Nonetheless, the irrational and inappropriate usage of antibiotics has resulted in rising trend of resistant organisms especially in critical care settings [5, 6]. Therefore, once the identity of the etiologic pathogen and the antimicrobial susceptibility data are available, every attempt should be made to narrow down the antibiotic spectrum. This is a critical component of antibiotic therapy through which a reduction in the cost, toxicity and development of antimicrobial resistance in the community can be accomplished.
The micro-organisms and their antibiotic susceptibility pattern vary among different healthcare facilities and geographical areas. The antibiograms provide a summary of in vitro activity of antimicrobials of an institution or geographical area. So, the decisions regarding initial antimicrobial therapy should be based on the institution’s specific antibiograms. Clinicians must choose empirical antibiotic therapy aimed at both maximizing outcomes and minimizing the emergence of resistance. With blood culture being one of the most reliable investigations for bacterial isolation and detection, we designed the present study to determine the bacterial profile of bloodstream infections (BSI) and their antibiotic susceptibility patterns among the clinically diagnosed cases of sepsis in patients presenting to our surgical intensive care unit (ICU) to direct the antibiotic treatment of hospital acquired infections in the ICU.
After approval from institutional ethical committee, this prospective study was conducted over a period of eighteen months in the 6- bedded surgical ICU of a tertiary care hospital. All patients of either sex between the age of 15-60 years, admitted to the ICU during the study period were included. Patients shifted to ICU for monitoring during postoperative period, mortality within 24 hours of admission and patients transferred to another speciality team were excluded. Written informed consent was obtained from either the patient or relatives of the included patients. The blood culture sample of these patients were collected when the patient presented with any two of the following four features,alongwitha suspected source of infection i.e. temperature>38$^{o}$C or90 beats/min, respiratory rate >24/min and Total leucocyte counts >12000/cu mm or < 4000/cu mm.
Collection of blood sample for blood culture was done using standard aseptic techniques. 10 ml of blood specimen was collected and inoculated into brain heart infusion (BHI) broth at the blood to broth ratio of 1:10. After incubation at 37$^{o}$C for 24 and 48hours, blind subcultures were made on Macconkey agar and blood agar plates (Hi Media Laboratories, India). After 24 hrs of aerobic incubation at 37$^{o}$C, the plates were observed for bacterial growth. Identification of significant isolates and their antimicrobial susceptibility tests was carried out as per Clinical and Laboratory Standards Institute (CLSI) guidelines, 2012 [7]. Antimicrobial sensitivity patterns of isolated organisms were identified by Kirby Bauer’s disc diffusion method on Mueller Hinton Agar media [8]. Interpretations of antibiotic susceptibility results were made according to the guidelines of interpretative zone diameters of CLSI [7]. Antibiotics that were tested in this study include Amoxycillin-sulbactam, Cefuroxime, ceftriaxone,cefoperazone-sulbactum,cefipime,cefazolin,ceftazidime,piperacillin-tazobactum,imepenem, meropenem, linezolid, clarithromycin, azithromycin, clindamycin, norfloxacin, ofloxacin, levofloxacin, sparfloxacin, gentamycin, amikacin, tobramycin, netilmycin, tigecycline, nitrofurantion, colistin, polymyxin-B, and vancomycin.
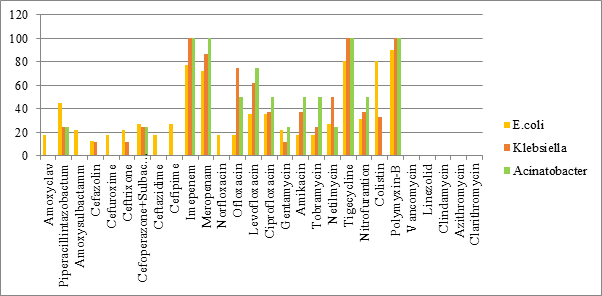
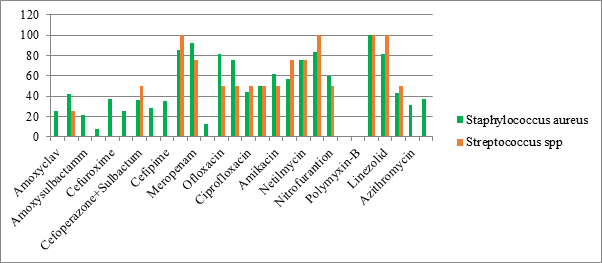
| Amoxyclav | Piperacillintazo | Amoxysulbactamm | Cefazolin | Cefuroxime | Ceftrixone | Cefoperazone+Sulbactum | Ceftazidime | Cefpime | Imepenem | Meropenam | Norfloxacin | Ofloxacin | Levofloxacin | ciprofloxacin | Gentamycin | Amikacin | tobramycin | netilmycin | Tigecycline | nitrofurantion | Colistin | Polymyxin-B | Vancomycin | Linezolid | Clindamycin | Azithromycin | Clarithromycin | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E.coli(11) | 18 | 45 | 22 | 13 | 18 | 22 | 27 | 18 | 27 | 77 | 72 | 18 | 18 | 36 | 36 | 22 | 18 | 18 | 27 | 81 | 31 | 81 | 90 | ||||||
| Staphylococcus aureus(8) | 25 | 42 | 21 | 8 | 37 | 25 | 36 | 28 | 35 | 85 | 92 | 13 | 81 | 75 | 44 | 50 | 62 | 57 | 75 | 83 | 60 | 100 | 81 | 43 | 31 | 37 | |||
| Klebsiella(4) | 0 | 25 | 0 | 12 | 0 | 12 | 25 | 0 | 0 | 100 | 87 | 0 | 75 | 62 | 37 | 12 | 37 | 25 | 50 | 100 | 37 | 33 | 100 | ||||||
| Streptococcus spp(2) | 0 | 25 | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 100 | 75 | 0 | 50 | 50 | 50 | 50 | 50 | 75 | 75 | 100 | 50 | 100 | 100 | 50 | |||||
| Acinetobacter(2) | 0 | 25 | 0 | 0 | 0 | 25 | 25 | 25 | 0 | 100 | 100 | 50 | 50 | 75 | 50 | 25 | 50 | 50 | 25 | 100 | 50 | 50 | 100 |
Nosocomial infections cause significant morbidity and mortality in patients admitted to ICUs worldwide. Antibiotics form a considerable portion of the immense economic burden borne by these patients [5]. However, inappropriate use of antibiotics may lead to antimicrobial resistance causing increasing mortality and healthcare costs. This study was undertaken to study the spectrum of the bacterial isolates causing blood stream infection in ICU patients and their antibiotic susceptibility pattern which could guide the formulation of antibiogram and future antibiotic policy.The Blood culture positivity rate in our study was observed to be 21.43% which was similar to the studies conducted by Alam et al., and others [10, 11, 12]. Though these were lower than the incidence observed by Parihar et al., and others [13,14, 15, 16, 17]. But, these blood culture rates were however higher as compared to a few other studies where the blood culture positive rates ranged only from 9.94% – 11.2% [18, 19, 20]. These differences in the positivity rates may be due to the difference in methodology used for blood culture, the study design, nature of patient population, epidemiological difference in etiological agents, geographical differences and differences in the infection control policies [19, 20, 21].
In our study, 60% of the infections were caused by GNBand36.67% of the infection was due to GPB. This finding was comparable to most of the studies from India and other developing countries, where Gram-negative bacteria have been reported to be the most common cause of bacteraemia in hospitalized patients [21, 22, 23]. In contrast, Arora et al., [11] and Shrestha et al., [17] have reported gram positive bacterial dominance in blood stream infections. Escherichia coli (36.67%) was the predominant Gram-negative isolate in our study, followed by Klebsiella (13.33%). These findings are in concordance with findings of Gupta et al., [23]. However, some other studies have isolated Pseudomonas and Acinetobacter predominantly [24]. This may be due to different antibiotic prescription policies. In GPB isolates, Staphylococcus aureus (26.67%) formed the majority followed by Streptococcus (6.67%). Similar findings were reported by Gupta et al., and Parihar et al., [23, 13], that Candida was seen in 3.33% of positive blood culture and all were non albicans Candida species.
Antibiotic resistance is a major concern in ICU worldwide and especially in India. Critical care areas are the major foci of antimicrobial resistance in hospitals [1]25,26}. Overuse of antibiotics is the leading cause of selection pressure on organisms and thereby, antimicrobial resistance [1]27}.
All the three major GNB isolates; E.coli, Klebsiella and Acinetobacter showed high degree of resistance to penicillins, cephalosporins and piperacillin-tazobactum. Susceptibility to levofloxacin, ofloxacin, amikacin and netilmycin ranged from intermediate to high in Klebsiella and Acinetobacter while E coli showed high resistance. Similar findings were noted by Parajuli et al., [17]. The increasing resistance to Colistin is a troublesome finding as it further decreases the antimicrobials available in our armamentarium for treatment of infections, especially multidrug resistant variants. On the positive side, all the three GNB isolates demonstrated higher susceptibility to Carbapenems, tigecycline and polymixin B. This is in contrast to studies [24, 25, 26, 27, 28, 29], which showed high level of resistance to Carbapenems. The low resistance to Carbapenems in our study could be attributed in part to the practice of administering the Carbapenems as infusions in our institute which has shown to limit the resistance to these antibiotics.
Both GPBisolates, staphylococcus and streptococcus showed high resistance to penicillins, cephalosporins and macrolides. The sensitivity for aminoglycosides, clindamycin and fluoroquinolones was intermediate with higher sensitivity noted to ofloxacin and levofloxacin. However, the sensitivity to Carbapenems, tigecyclin, linezolid and vancomycin was as high as upto100%. The high degree of resistance to $\beta$-lactams, most cephalosporins, and increasing resistance to fluoroquinolones and aminoglycosides among both GPB and GNB in our study was also established in many other studies, see [11, 27, 28, 29]. This is probably because these are the most frequently prescribed antibiotics in developing nations. Another frequently observedissue in developing nations is the easy availability of antibiotics as over the counter preparations.